Version 5 of Biological Mutation Rate and TCL demo example calculator, numerical analysis
Updated 2021-05-10 19:32:07 by goldStarter Wiki Page
This page is under development. Comments are welcome, but please load any comments in the comments section at the bottom of the page. Please include your wiki MONIKER and date in your comment with the same courtesy that I will give you. Aside from your courtesy, your wiki MONIKER and date as a signature and minimal good faith of any internet post are the rules of this TCL-WIKI. Its very hard to reply reasonably without some background of the correspondent on his WIKI bio page. Thanks, gold 12Jan2019
Title: Biological Mutation Rate and TCL demo example calculator, numerical analysis
- Appendix Code
- Hidden Comments Section
Preface
gold12Jan2019. Here are some calculations using TCL expressions on biological mutation rate.
Introduction
The first testcase was published by Dr Sene Carroll, HHMI. The mutation rate is 1 in 25E6. The offspring equals 5000 females times 5 offspring per year or 25000 offspring per year. The mutation should occur 25E6/25000 or every 1000 years. The geologic interval was 1.7E6 years. Mutation would occur 1.7E6/1000 or 1700 times over the geologic interval.
Calculator Usage
As as alternate way of using the calculator
Expressions into TCL.
Testcases are important in development of TCL programs.
Conclusions
By inspection
Table 1: Tables
Table 2: Tables
Testcases Section
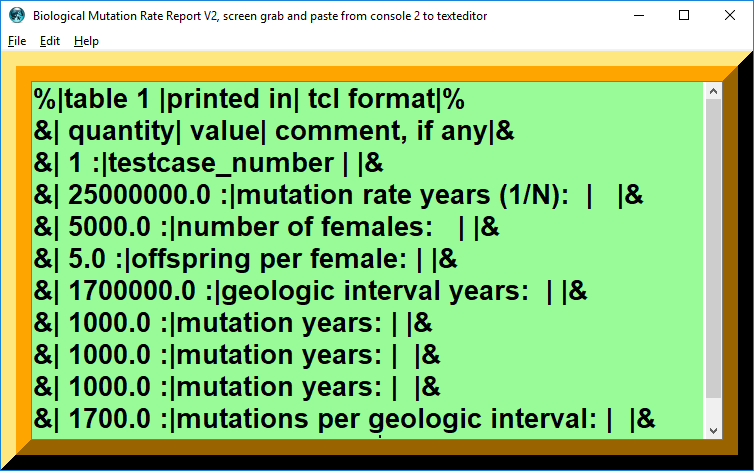
Testcase 1
| table 1 | printed in | tcl format |
|---|---|---|
| quantity | value | comment, if any |
| 1 : | testcase_number | |
| 25000000.0 : | mutation rate years (1/N): | |
| 5000.0 : | number of females: | |
| 5.0 : | offspring per female: | |
| 1700000.0 : | geologic interval years: | |
| 1000.0 : | mutation years: | |
| 1000.0 : | mutation years: | |
| 1000.0 : | mutation years: | |
| 1700.0 : | mutations per geologic interval: |
Testcase 2
| table 2 | printed in | tcl format |
|---|---|---|
| quantity | value | comment, if any |
| 2 : | testcase_number | |
| 20000000.0 : | mutation rate years (1/N): | |
| 7000.0 : | number of females: | |
| 5.0 : | offspring per female: | |
| 600000.0 : | geologic interval years: | |
| 1000.0 : | mutation years: | |
| 571.428 : | mutation years: | |
| 571.428 : | mutation years: | |
| 1050.0 : | mutations per geologic interval: |
Testcase 3
| table 3 | printed in | tcl format |
|---|---|---|
| quantity | value | comment, if any |
| 3 : | testcase_number | |
| 10000000.0 : | mutation rate years (1/N): | |
| 9000.0 : | number of females: | |
| 5.0 : | offspring per female: | |
| 800000.0 : | geologic interval years: | |
| 1000.0 : | mutation years: | |
| 222.222 : | mutation years: | |
| 222.222 : | mutation years: | |
| 3600.0 : | mutations per geologic interval: |
Screenshots Section
figure 1. Calculator Screenshot
Biological Mutation Rate and TCL calculator screenshot
figure 2. Console Screenshot
References:
- google search engine < Rock pocket mice >
- Wikipedia search engine < hardy-weinberg >
- Wikipedia search engine < Sene Carroll >
- startpage search engine <Sene Carroll >
- TCL-WIKI search engine < biology >
- On S.N. Bernstein’s derivation of Mendel’s Law
- and ‘rediscovery’ of the Hardy-Weinberg distribution
- Alan Stark and Eugene Seneta, University of Sydney
- Rock Pocket Mice, University of Utah
- Hardy-Weinberg Equilibrium Model, by Dennis O'Neil
- Albinism: a sample hardy-weinberg problem,
- by Dennis O'Neil
- Allele and phenotype frequencies
- in rock pocket mouse populations,hhmi
- Mendelian Proportions in a Mixed Population, Hardy,
- Trinity College, 1908
- Wikipedia, Hardy-Weinberg principle
- Wikipedia, Wahlund effect
- Wikipedia, Multinomial distribution
- Hardy-Weinberg Principle of Equilibrium
- UC Davis Library, the California State University
Pseudocode & Equations Section
more than one formula for 1) calculator shell
Appendix Code
Trial Code
# TCL source code follows
# pretty print from autoindent and ased editor
# Biological Mutation Rate calculator V2
# written on Windows XP on TCL
# working under TCL version 8.6
# gold on TCL Club, 12Dec2018
package require Tk
package require math::numtheory
namespace path {::tcl::mathop ::tcl::mathfunc math::numtheory }
set tcl_precision 17
frame .frame -relief flat -bg aquamarine4
pack .frame -side top -fill y -anchor center
set names {{} {mutation rate years (1/N) :} }
lappend names {number of females:}
lappend names {offspring per female: }
lappend names {geologic interval years : }
lappend names {mutation years:}
lappend names {mutation years: }
lappend names {mutation years: }
lappend names {mutations per geologic interval: }
foreach i {1 2 3 4 5 6 7 8} {
label .frame.label$i -text [lindex $names $i] -anchor e
entry .frame.entry$i -width 35 -textvariable side$i
grid .frame.label$i .frame.entry$i -sticky ew -pady 2 -padx 1 }
proc about {} {
set msg "Calculator for Biological Mutation Rate
from TCL
# gold on TCL Club, 12Dec2018 "
tk_messageBox -title "About" -message $msg }
proc self_help {} {
set msg " Biological Mutation Rate V2
from TCL ,
# self help listing
# problem, Biological Mutation Rate V2
# Recommended procedure is push
# testcase and fill frame,
# change first three entries etc, push solve,
# and then push report. Report allows copy and paste
# from console to conventional texteditor.
# For testcases, th
# testcase number is internal to the calculator and
# will not be printed until the report button is pushed
# for the current result numbers.
# >>> copyright notice <<<
# This posting, screenshots, and TCL source code is
# copyrighted under the TCL/TK license terms.
# Editorial rights and disclaimers
# retained under the TCL/TK license terms
# and will be defended as necessary in court.
Conventional text editor formulas or
grabbed from internet
screens can be pasted into green console.
# gold on TCL Club, 12Dec2018 "
tk_messageBox -title "Self_Help" -message $msg }
proc calculate { } {
global answer2
global side1 side2 side3 side4 side5
global side6 side7 side8
global testcase_number
incr testcase_number
set side1 [* $side1 1. ]
set side2 [* $side2 1. ]
set side3 [* $side3 1. ]
set side4 [* $side4 1. ]
set side5 [* $side5 1. ]
set side6 [* $side6 1. ]
set side7 [* $side7 1. ]
set side8 [* $side8 1. ]
set generation_interval_years $side1
set females $side2
set offspring $side3
set geologic_interval $side4
set offspring_over_interval [* $females $offspring ]
set mutation_instance [ expr { (1.0 * $generation_interval_years)/( $females * $offspring ) } ]
set mutation_instance2 [ expr { (1.0 * $geologic_interval)/ $mutation_instance } ]
set side6 $mutation_instance
set side7 $mutation_instance
set side8 $mutation_instance2
}
proc fillup {aa bb cc dd ee ff gg hh} {
.frame.entry1 insert 0 "$aa"
.frame.entry2 insert 0 "$bb"
.frame.entry3 insert 0 "$cc"
.frame.entry4 insert 0 "$dd"
.frame.entry5 insert 0 "$ee"
.frame.entry6 insert 0 "$ff"
.frame.entry7 insert 0 "$gg"
.frame.entry8 insert 0 "$hh"
}
proc clearx {} {
foreach i {1 2 3 4 5 6 7 8 } {
.frame.entry$i delete 0 end } }
proc reportx {} {
global answer2
global side1 side2 side3 side4 side5
global side6 side7 side8
global testcase_number
global wavelength wavelength2
global wavelength6 wavelength10
global wavelengthsq surfacearea
global megafrequency
console eval {.console config -bg palegreen}
console eval {.console config -font {fixed 20 bold}}
console eval {wm geometry . 40x20}
console eval {wm title . " Biological Mutation Rate Report V2, screen grab and paste from console 2 to texteditor"}
console eval {. configure -background orange -highlightcolor brown -relief raised -border 30}
console show;
puts "%|table $testcase_number |printed in| tcl format|% "
puts "&| quantity| value| comment, if any|& "
puts "&| $testcase_number :|testcase_number | |&"
puts "&| $side1 :|mutation rate years (1/N): | |&"
puts "&| $side2 :|number of females: | |& "
puts "&| $side3 :|offspring per female: | |& "
puts "&| $side4 :|geologic interval years: | |&"
puts "&| $side5 :|mutation years: | |&"
puts "&| $side6 :|mutation years: | |&"
puts "&| $side7 :|mutation years: | |&"
puts "&| $side8 :|mutations per geologic interval: | |&"
}
frame .buttons -bg aquamarine4
::ttk::button .calculator -text "Solve" -command { set side8 0 ; calculate }
::ttk::button .test2 -text "Testcase1" -command {clearx;fillup 25E6 5000. 5. 1.7E6 1000. 1000. 1000. 1700. }
::ttk::button .test3 -text "Testcase2" -command {clearx;fillup 20E6 7000. 5. 0.6E6 1000. 570. 570. 1050. }
::ttk::button .test4 -text "Testcase3" -command {clearx;fillup 10E6 9000. 5. 0.8E6 1000. 222. 222. 3600. }
::ttk::button .clearallx -text clear -command {clearx }
::ttk::button .about -text about -command {about}
::ttk::button .self_help -text self_help -command { self_help }
::ttk::button .cons -text report -command { reportx }
::ttk::button .exit -text exit -command {exit}
pack .calculator -in .buttons -side top -padx 10 -pady 5
pack .clearallx .cons .self_help .about .exit .test4 .test3 .test2 -side bottom -in .buttons
grid .frame .buttons -sticky ns -pady {0 10}
. configure -background aquamarine4 -highlightcolor brown -relief raised -border 30
wm title . "Biological Mutation Rate Calculator V2"Hidden Comments Section
Please include your wiki MONIKER and date in your comment with the same courtesy that I will give you. Thanks, gold 12Jan2019
| Category Numerical Analysis | Category Toys | Category Calculator | Category Mathematics | Category Example | Toys and Games | Category Games | Category Application | Category GUI |